how to send a 3d pharmacophore search query.
Published 14 years ago • 850 plays • Length 0:33Download video MP4
Download video MP3
Similar videos
-
 1:40
1:40
how to extract a 3d pharmacophore from a ligand.
-
 0:50
0:50
how to search a 2d pharmacophore.
-
 1:18
1:18
how to edit a 3d pharmacophore.
-
 7:12
7:12
how to: run the pharmacophore search using csd-crossminer
-
 0:34
0:34
how to draw a chemical search query.
-
 14:17
14:17
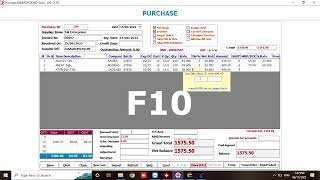
medisoft ® - how to import or manually create a new purchase bill
-
 4:56
4:56
3d printing of nutraceuticals & pharmaceuticals
-
 35:26
35:26
molecular docking for beginners | autodock full tutorial | bioinformatics
-
 0:18
0:18
how to color a 2d chemical sketch by pharmacophore feature.
-
 1:30
1:30
introducing molscreen from molsoft.
-
 4:42
4:42
virtual ligand screening - part one
-
 10:02
10:02
core replacement and linker search in the icm 3d ligand editor
-
 11:08
11:08
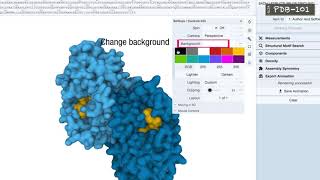
exploring pdb structures in 3d with molstar (mol*): introductory guide
-
 41:08
41:08
molcart giga search webinar
-
 39:01
39:01
molsoft webinar: ligand based lead discovery using atomic property fields
-
 1:47
1:47
how to convert a pdb file into an icm object.
-
 2:11
2:11
how to dock to two molecules.
-
 21:00
21:00
new molsoft bioisoster databases and datamining tools
-
 1:55
1:55
molsoft molcalcprop in knime
-
 49:57
49:57
rapid isostere discovery engine - ride